Atlas-based Segmentation of Deep Brain Structures

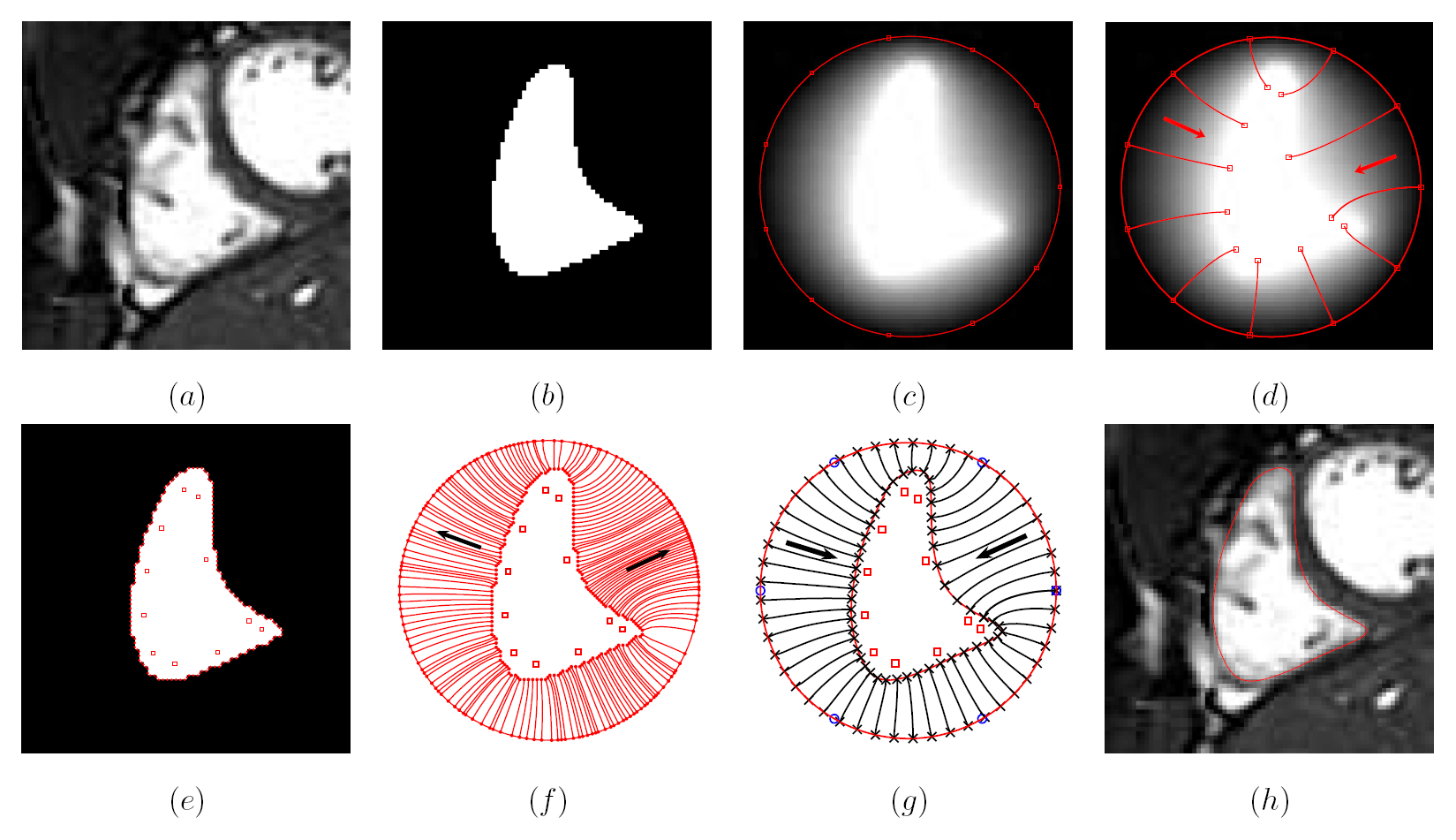
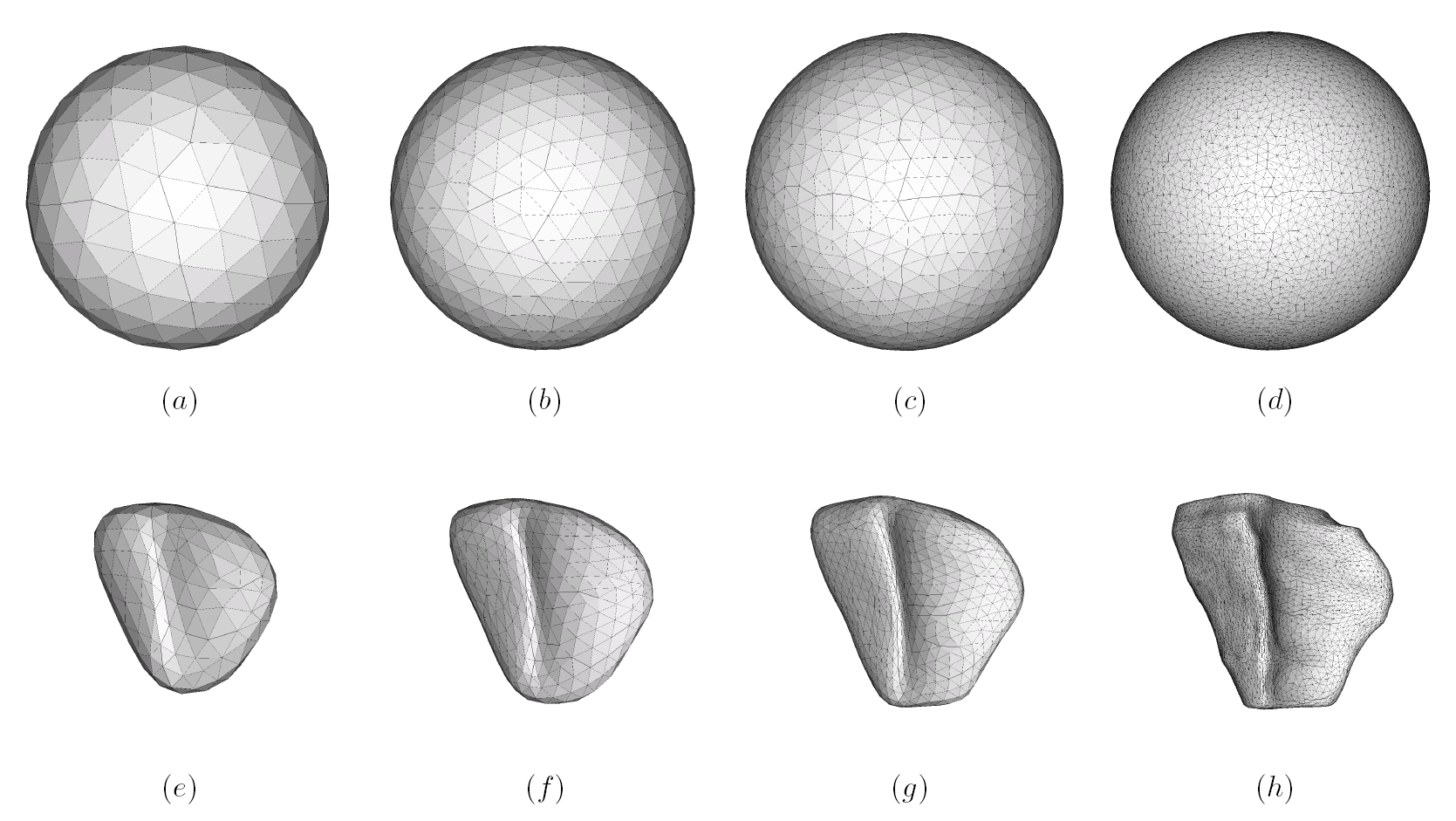
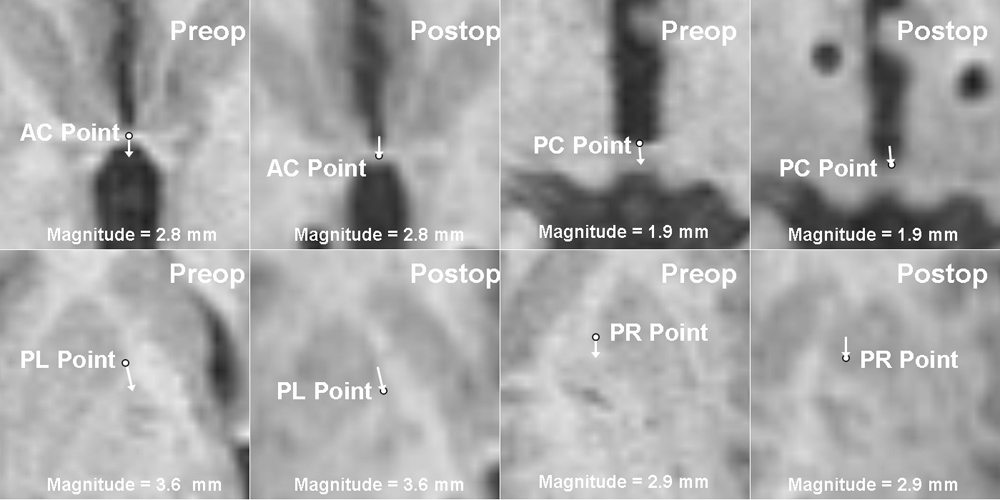
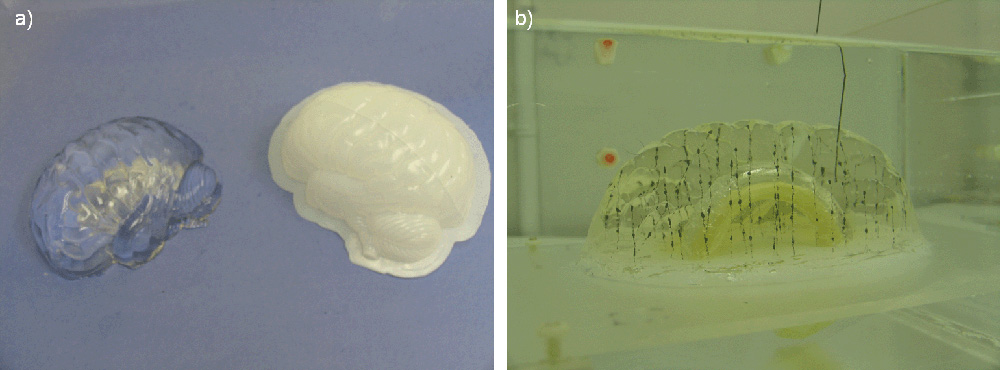
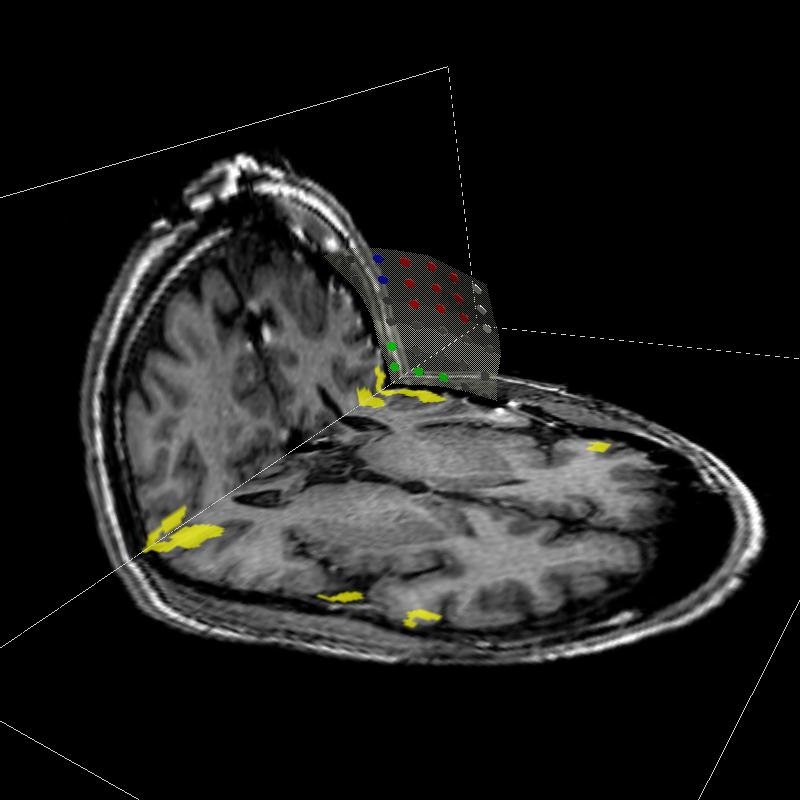
Deep brain structures are frequently used as targets in neurosurgical procedures. However, the boundaries of these structures are often not visible in clinically used MR and CT images. Techniques based on anatomical atlases and indirect targeting are used to infer the location of these targets intraoperatively. Initial errors of such approaches may be up to a few millimeters, which is not negligible. E.g. subthalamic nucleus is approximately 4x6 mm in the axial plane and the diameter of globus pallidus internus is approximately 8 mm, both of which are used as targets in deep brain stimulation surgery. To increase the initial localization accuracy of deep brain structures we have developed an atlas-based segmentation method that can be used for the surgery planning [1]. The atlas is a high resolution MR head scan of a healthy volunteer with nine deep brain structures manually segmented (Fig. 1). The quality of the atlas image allowed for the segmentation of the deep brain structures, which is not possible from the clinical MR head scans of patients.
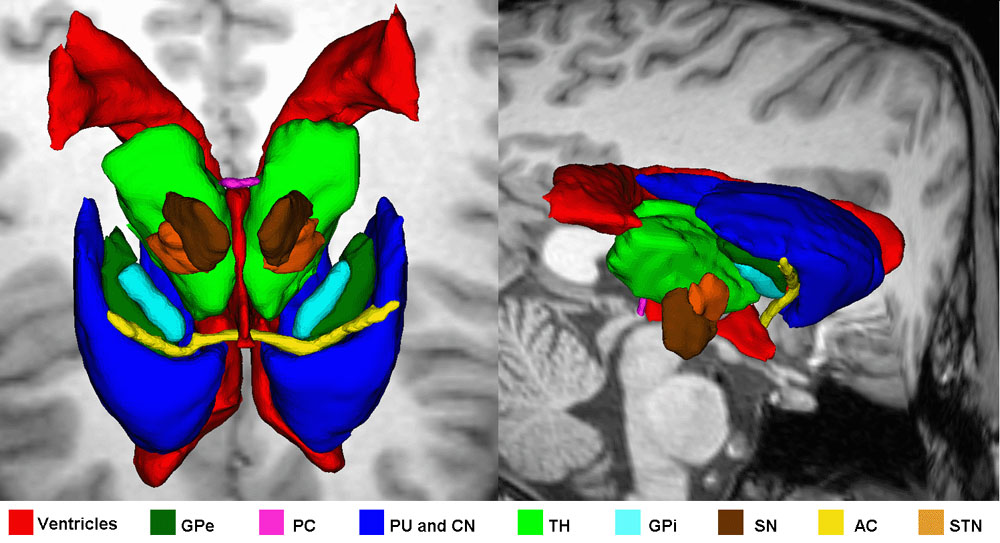

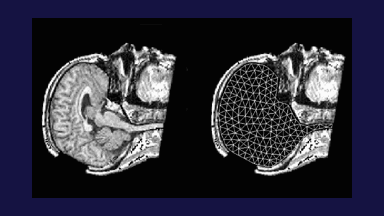
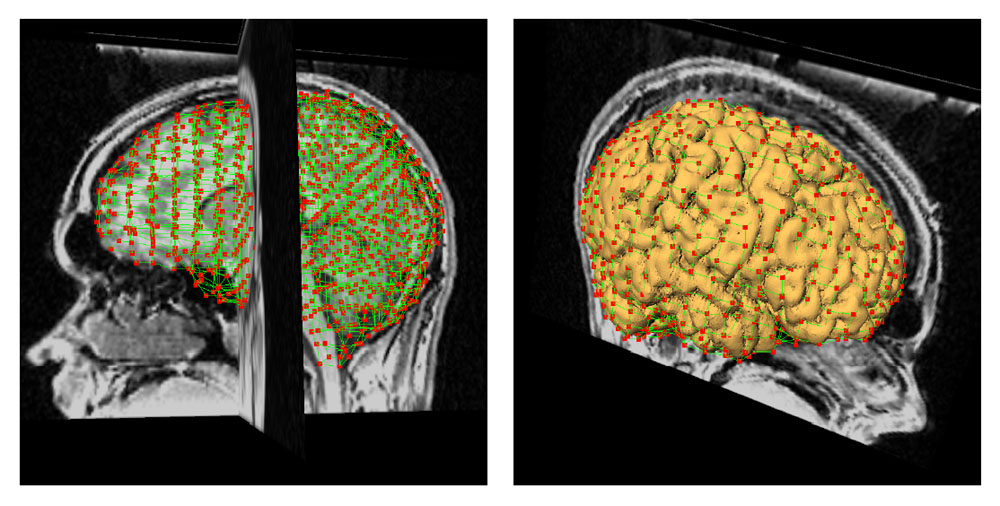
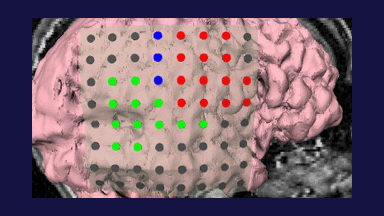
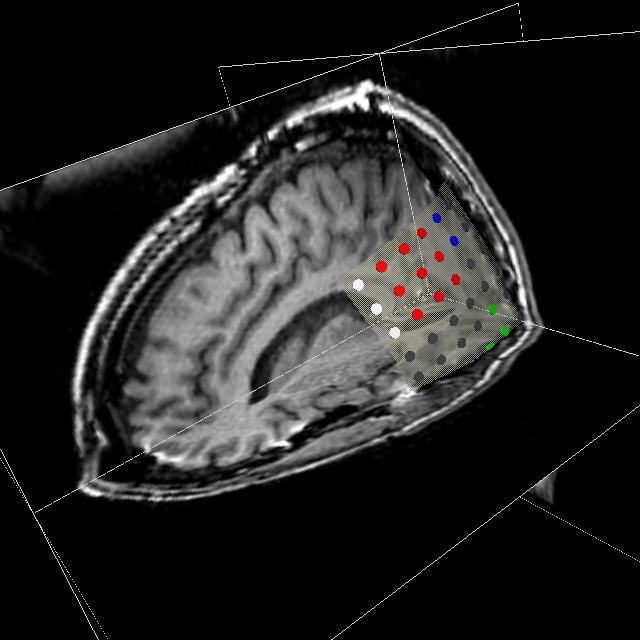
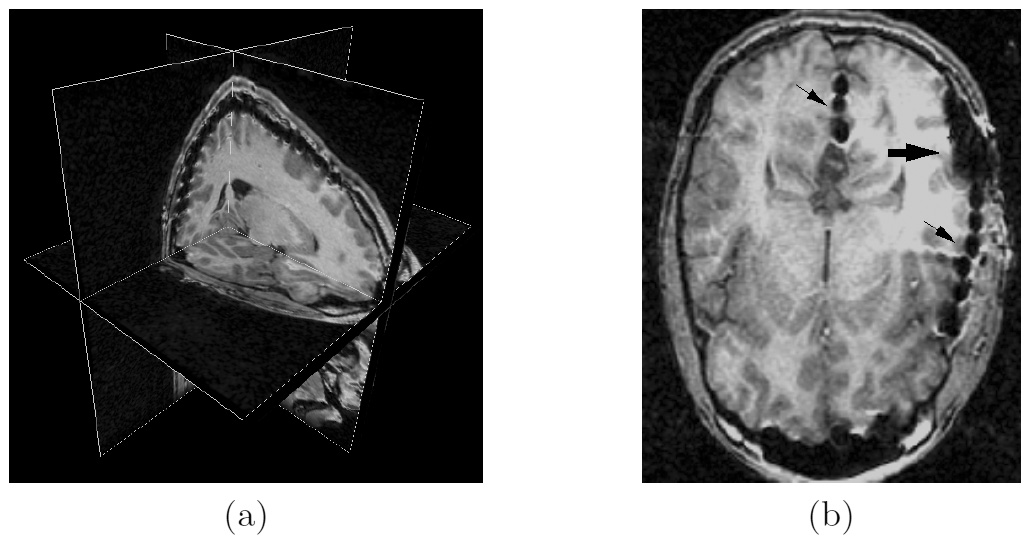
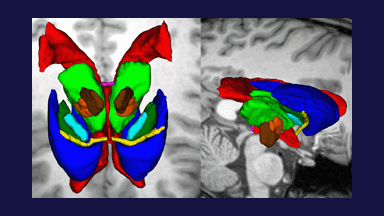
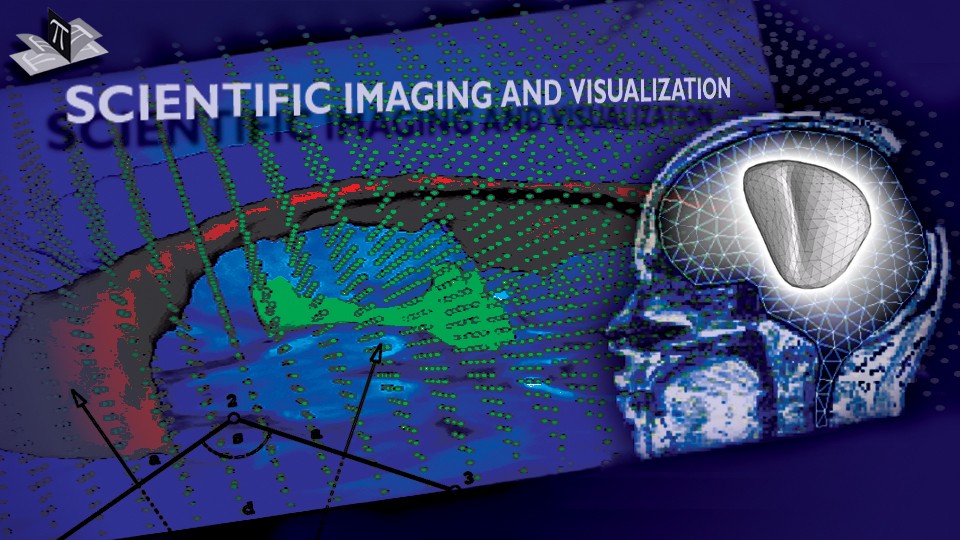
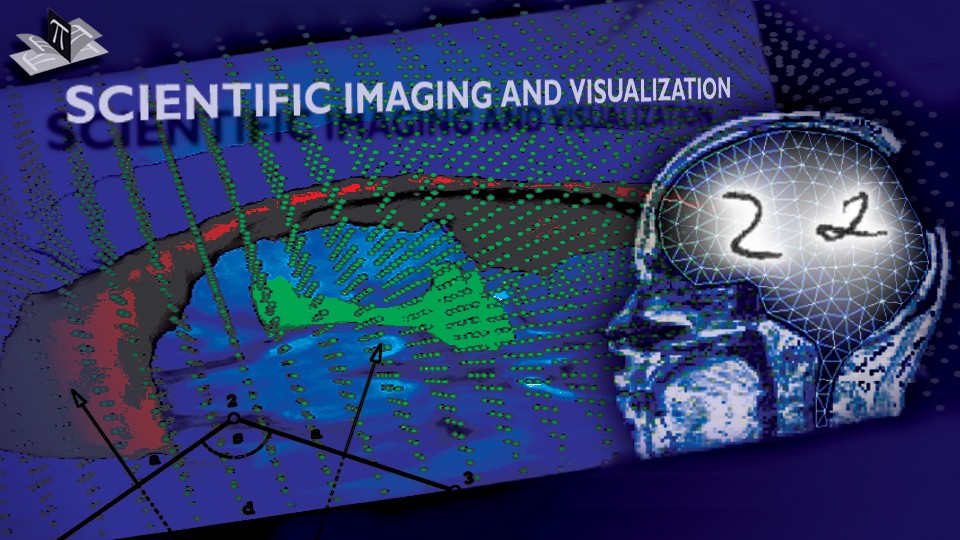
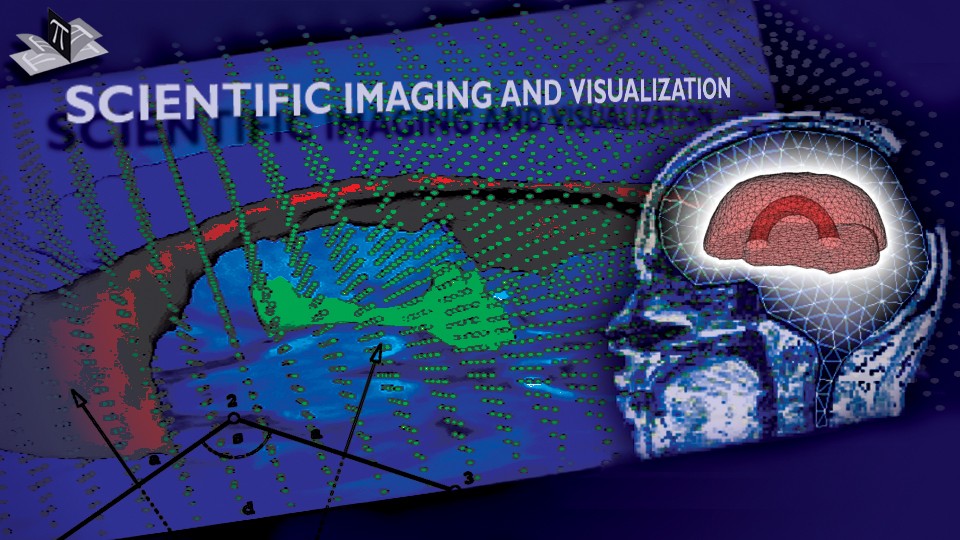
Figure 1: 3D models of the segmented deep brain structures of the atlas: inferior view with an axial image slice (left) and oblique view with three orthogonal image slices (right). The abbreviations are: putamen (PU), caudate nucleus (CN), globus pallidus internus (GPi), globus pallidus externus (GPe), anterior commissure (AC), posterior commissure (PC), thalamus (TH), subthalamic nucleus (STN), and substantia nigra (SN). The figure is from [1] and it is used with permission; Copyright © 2008 Society of Photo-Optical Instrumentation Engineers (SPIE). All rights reserved.
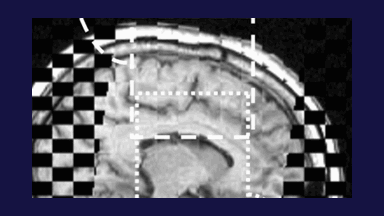
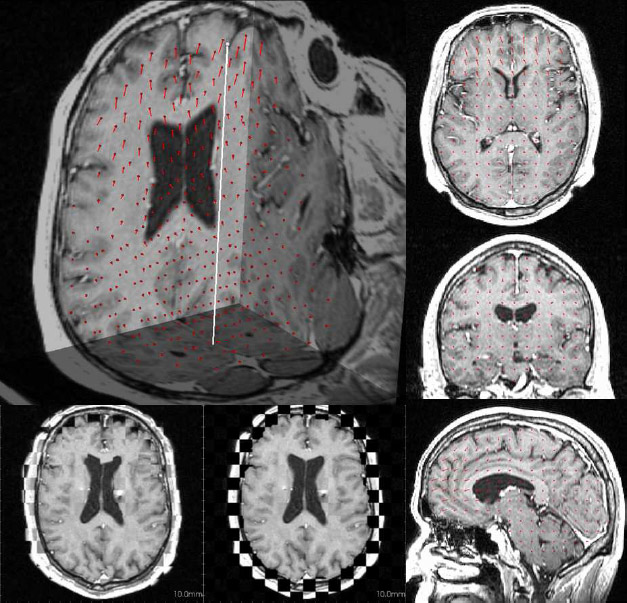
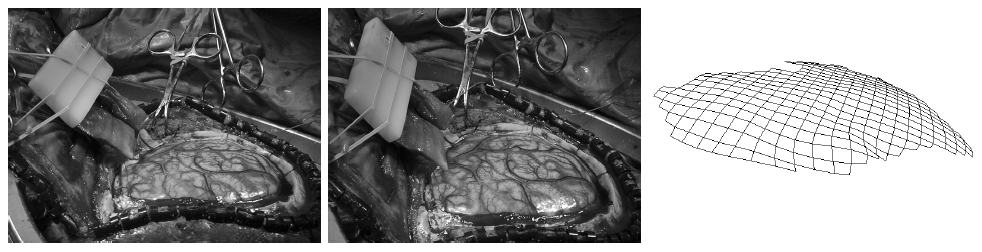
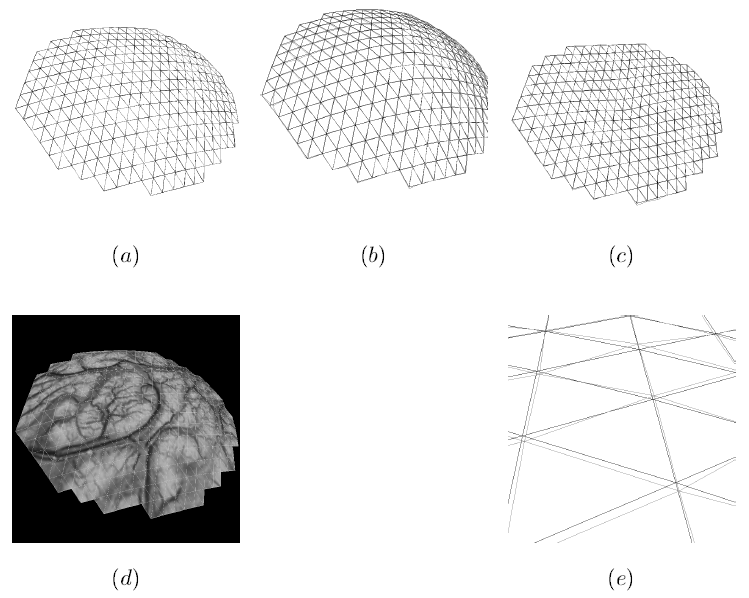
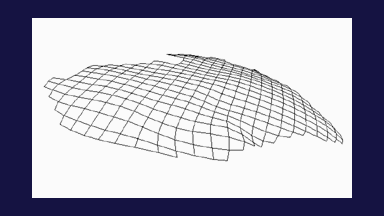
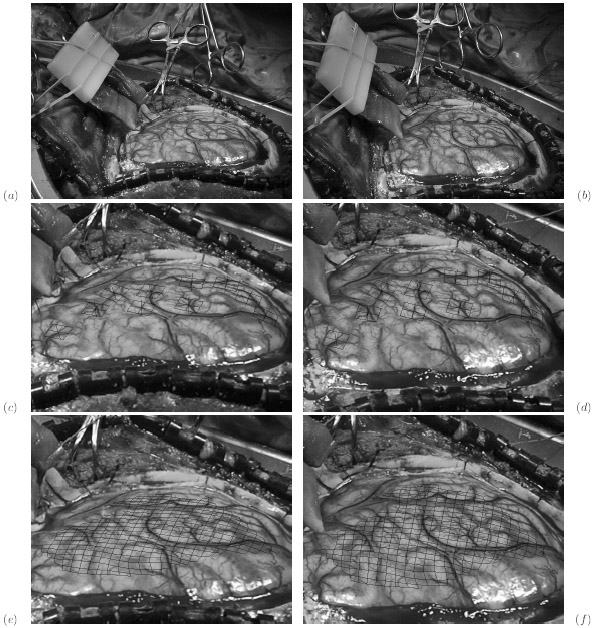
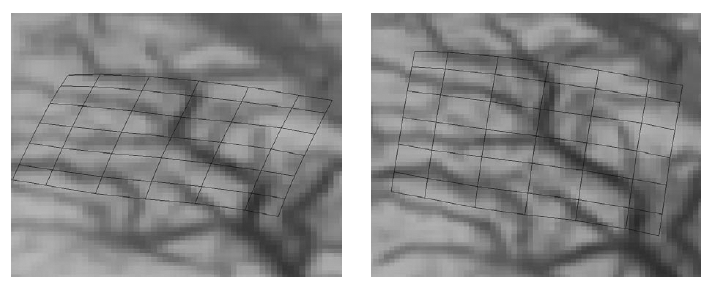
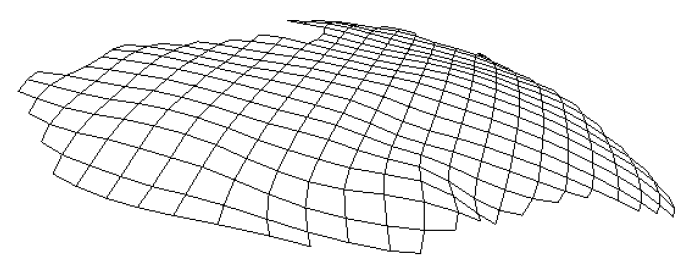
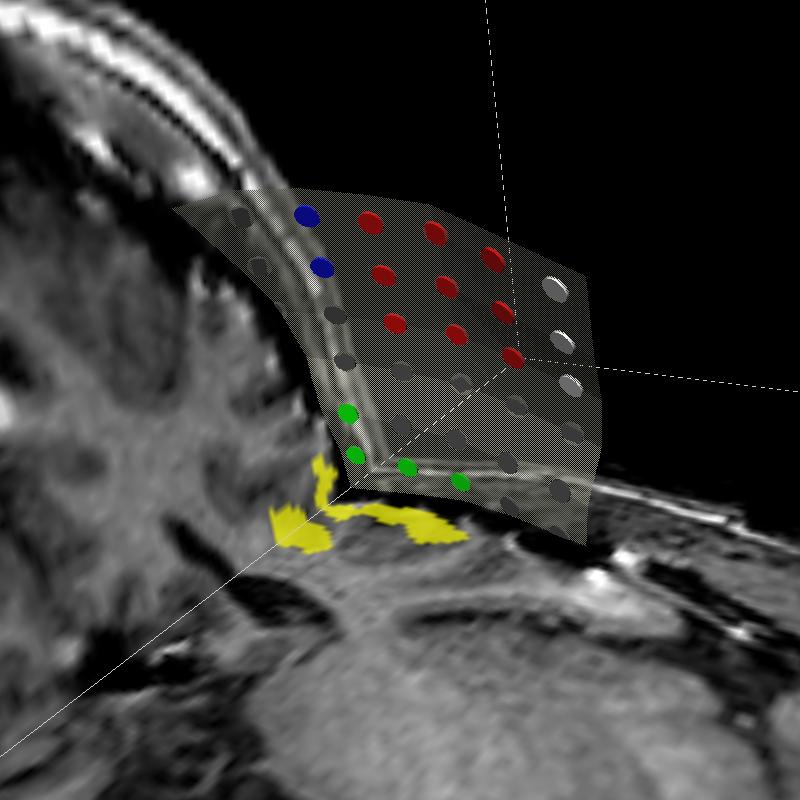
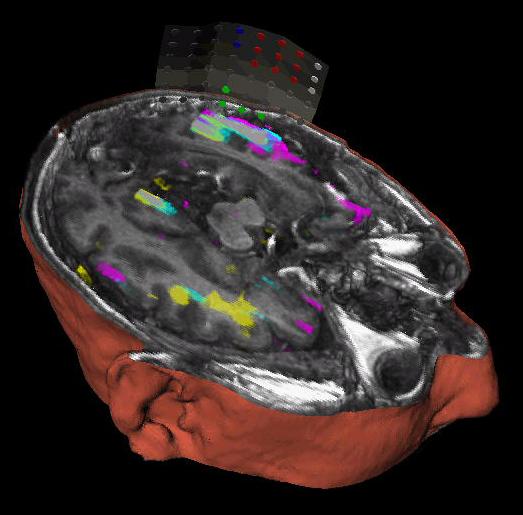
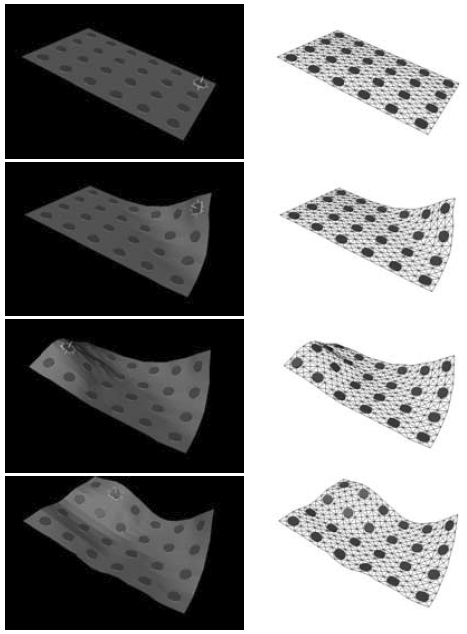
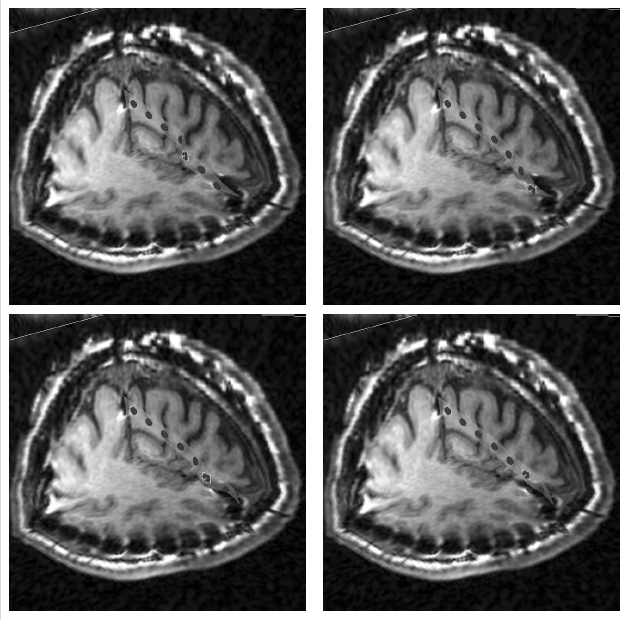
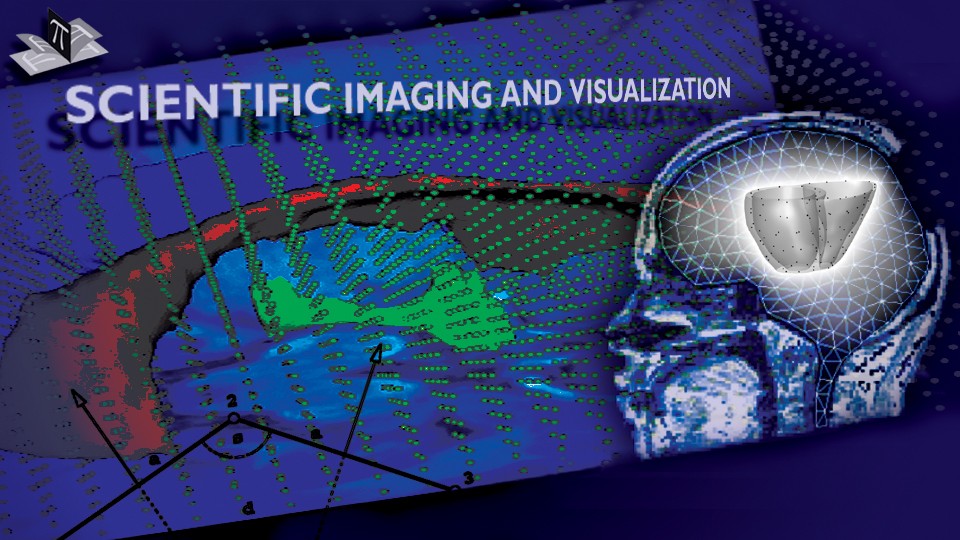
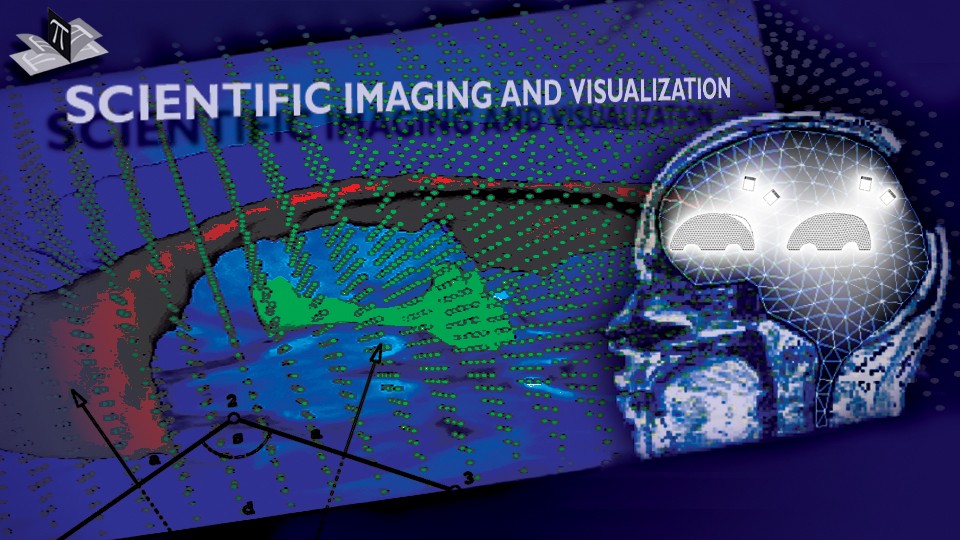
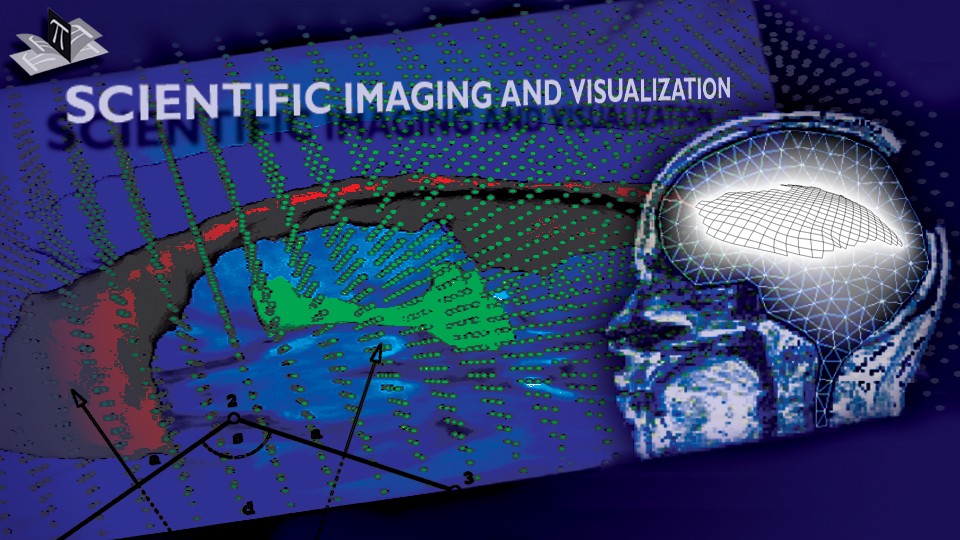
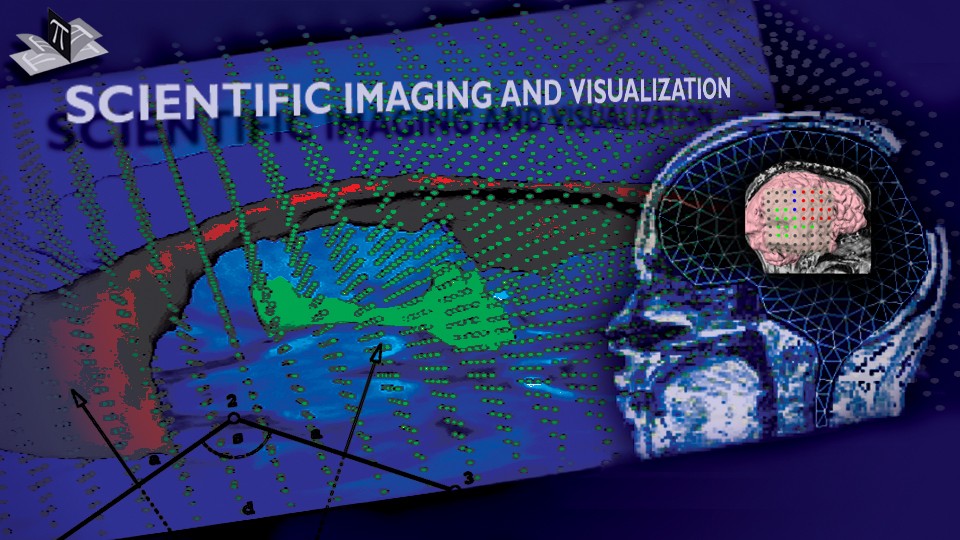
The subject image is non-rigidly registered to the atlas image using a two stage method. Results of the registration are shown in Figs. 2 and 3. The obtained transformation is used to map the segmented structures from the atlas to the subject image (Fig 4). We tested the approach on five subjects. The quality of the atlas-based segmentation was evaluated by visual inspection of the third and lateral ventricles, putamena, and caudate nuclei, which are visible in the subject MR images. The agreement of these structures for the five tested subjects was approximately 1 to 2 mm.
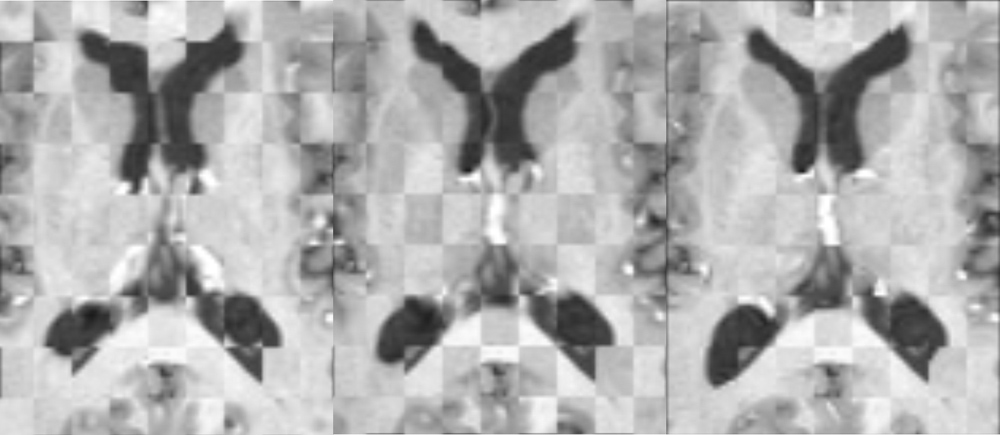

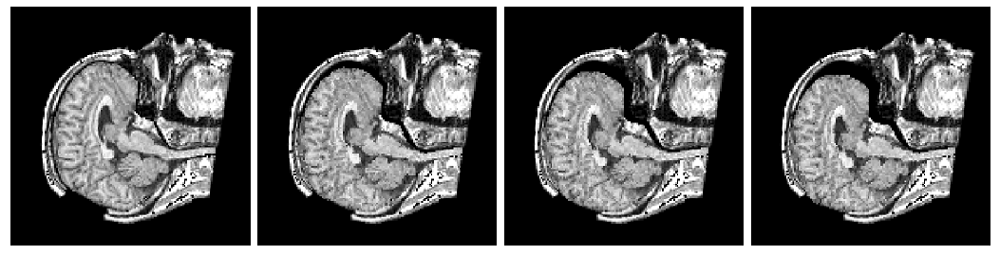
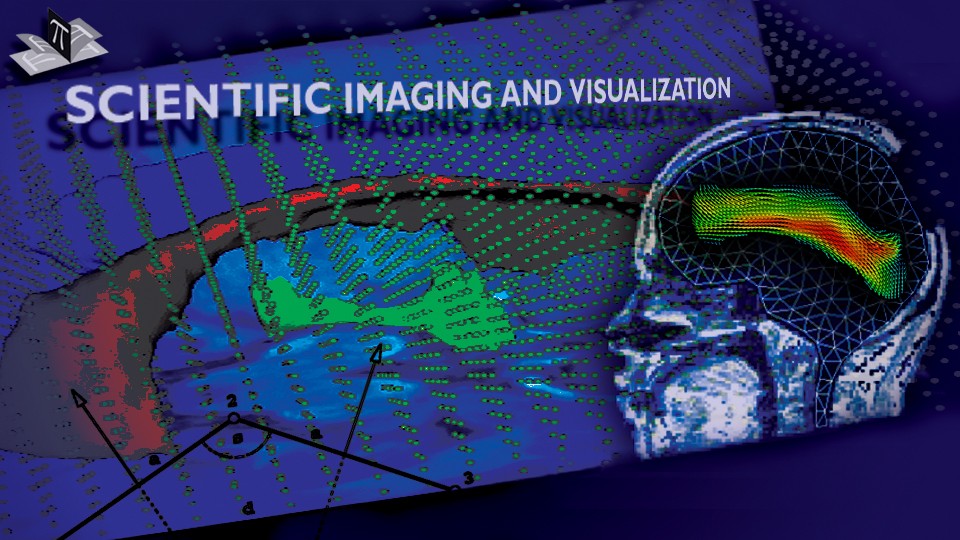
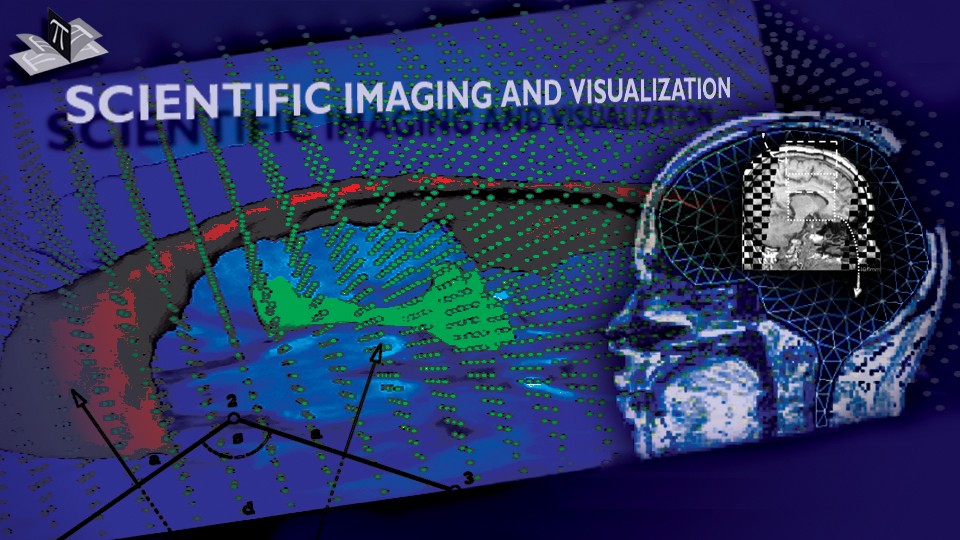
Figure 2: Axial checkerboard slices of the atlas image and a registered subject image after the initial affine alignment (left), Stage-1 of registration (center) and the final Stage-2 registration (right). Note the progressive improvement of alignment of the ventricles, PU and CN from initialization to the final registration. The figure is from [1] and it is used with permission; Copyright © 2008 Society of Photo-Optical Instrumentation Engineers (SPIE). All rights reserved.
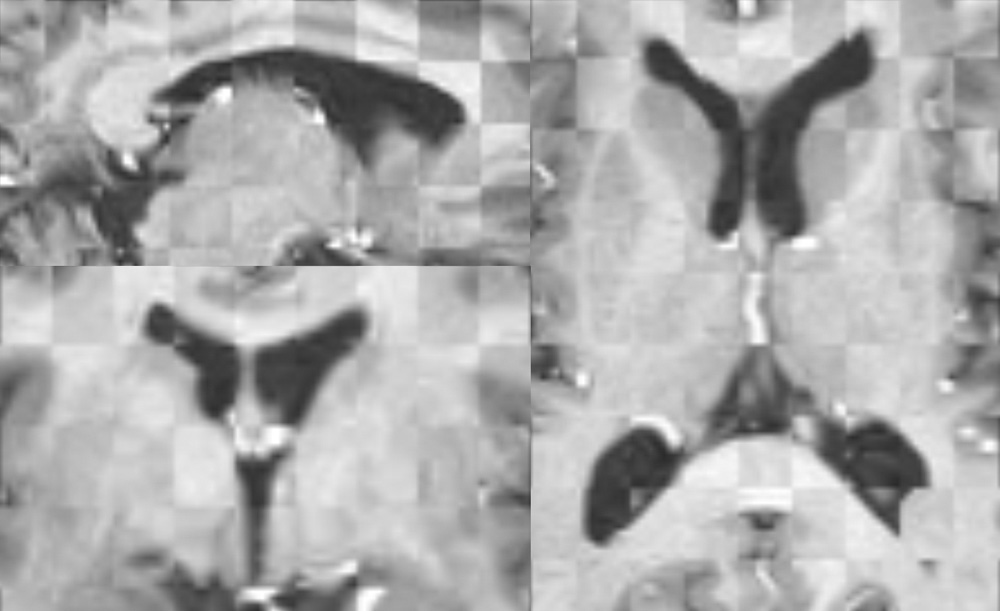

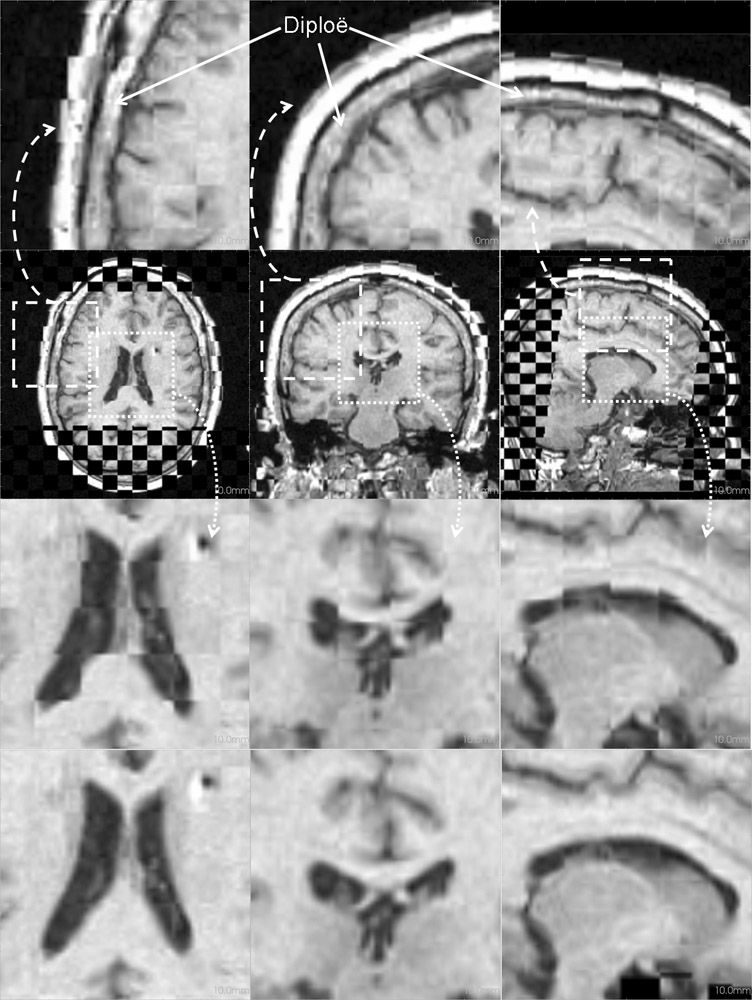
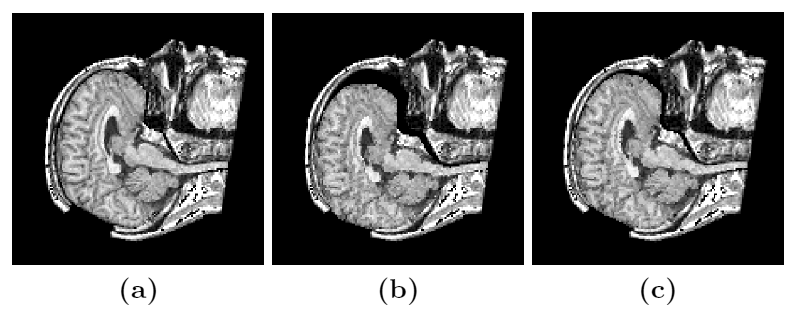
Figure 3: Axial (right), sagittal (top left) and coronal (bottom left) checkerboard slices of the atlas image and non-rigidly registered subject image. Note the good alignment of the ventricles, PU and CN at the boundaries of the checkerboards. Some apparent variations are because of difference in the intensities and protocols of the atlas and the subject image. The figure is from [1] and it is used with permission; Copyright © 2008 Society of Photo-Optical Instrumentation Engineers (SPIE). All rights reserved.
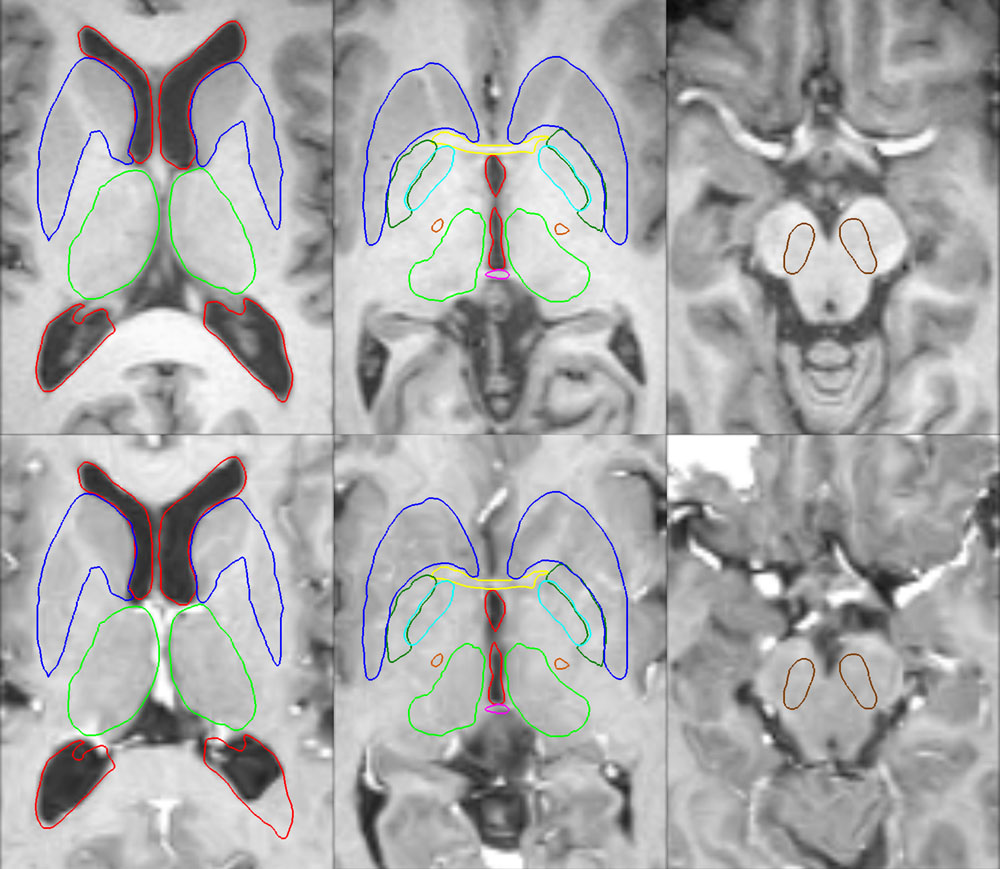

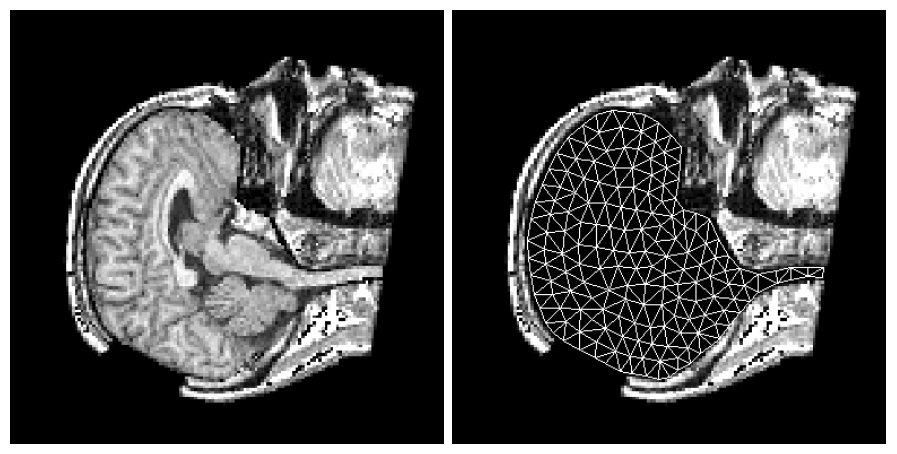
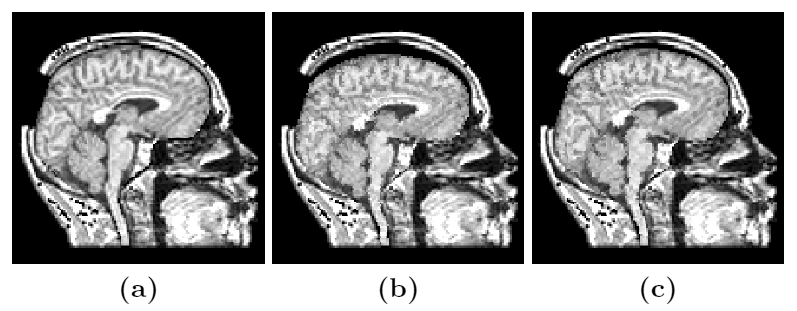
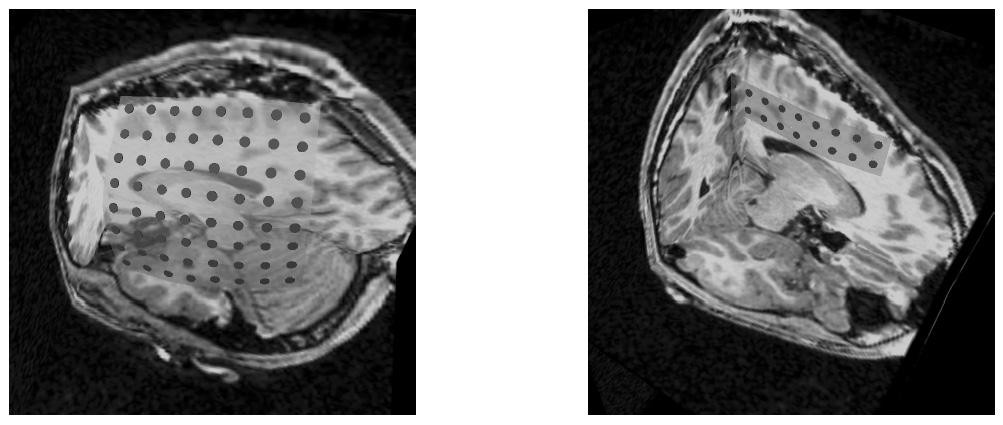
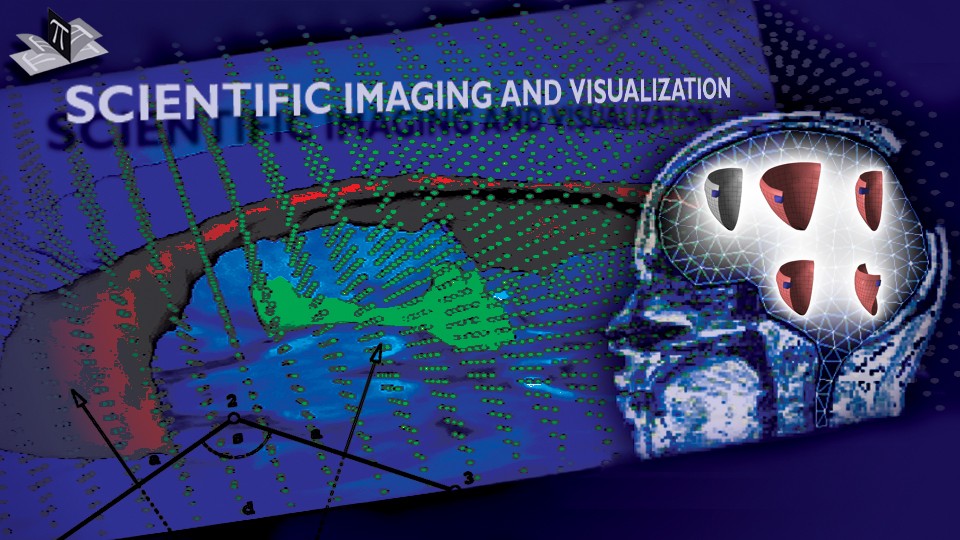
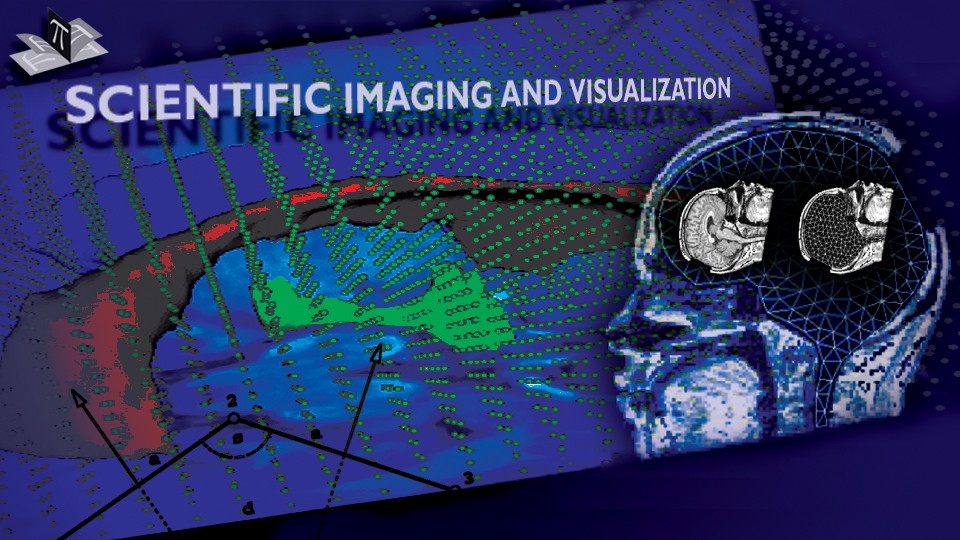
Figure 4: Deep brain region axial slices of the atlas image (top row) and a registered subject (bottom row) overlaid with the contours of the manually segmented atlas deep brain structures. The color coding is the same as in Fig. 1. The misalignment of the posterior part of the right lateral ventricle (bottom row, left) is due to a significant lateral ventricle shape difference between the subject and the atlas. The figure is from [1] and it is used with permission; Copyright © 2008 Society of Photo-Optical Instrumentation Engineers (SPIE). All rights reserved.
References:
[1] Khan, M., Mewes, K., Gross, E. R., Skrinjar, O., "Atlas-based Segmentation of Deep Brain Structures using Non-Rigid Registration", SPIE Medical Imaging 2008, San Diego, CA, USA, Vol. 6918, February 2008. LINK